Hyperspectral Image Segmentation
Hyperspectral image segmentation is the process of partitioning hyperspectral images into meaningful regions or objects.
Papers and Code
Scalable neural pushbroom architectures for real-time denoising of hyperspectral images onboard satellites
Jan 08, 2026The next generation of Earth observation satellites will seek to deploy intelligent models directly onboard the payload in order to minimize the latency incurred by the transmission and processing chain of the ground segment, for time-critical applications. Designing neural architectures for onboard execution, particularly for satellite-based hyperspectral imagers, poses novel challenges due to the unique constraints of this environment and imaging system that are largely unexplored by the traditional computer vision literature. In this paper, we show that this setting requires addressing three competing objectives, namely high-quality inference with low complexity, dynamic power scalability and fault tolerance. We focus on the problem of hyperspectral image denoising, which is a critical task to enable effective downstream inference, and highlights the constraints of the onboard processing scenario. We propose a neural network design that addresses the three aforementioned objectives with several novel contributions. In particular, we propose a mixture of denoisers that can be resilient to radiation-induced faults as well as allowing for time-varying power scaling. Moreover, each denoiser employs an innovative architecture where an image is processed line-by-line in a causal way, with a memory of past lines, in order to match the acquisition process of pushbroom hyperspectral sensors and greatly limit memory requirements. We show that the proposed architecture can run in real-time, i.e., process one line in the time it takes to acquire the next one, on low-power hardware and provide competitive denoising quality with respect to significantly more complex state-of-the-art models. We also show that the power scalability and fault tolerance objectives provide a design space with multiple tradeoffs between those properties and denoising quality.
Deep Global Clustering for Hyperspectral Image Segmentation: Concepts, Applications, and Open Challenges
Dec 30, 2025Hyperspectral imaging (HSI) analysis faces computational bottlenecks due to massive data volumes that exceed available memory. While foundation models pre-trained on large remote sensing datasets show promise, their learned representations often fail to transfer to domain-specific applications like close-range agricultural monitoring where spectral signatures, spatial scales, and semantic targets differ fundamentally. This report presents Deep Global Clustering (DGC), a conceptual framework for memory-efficient HSI segmentation that learns global clustering structure from local patch observations without pre-training. DGC operates on small patches with overlapping regions to enforce consistency, enabling training in under 30 minutes on consumer hardware while maintaining constant memory usage. On a leaf disease dataset, DGC achieves background-tissue separation (mean IoU 0.925) and demonstrates unsupervised disease detection through navigable semantic granularity. However, the framework suffers from optimization instability rooted in multi-objective loss balancing: meaningful representations emerge rapidly but degrade due to cluster over-merging in feature space. We position this work as intellectual scaffolding - the design philosophy has merit, but stable implementation requires principled approaches to dynamic loss balancing. Code and data are available at https://github.com/b05611038/HSI_global_clustering.
HyperCOD: The First Challenging Benchmark and Baseline for Hyperspectral Camouflaged Object Detection
Jan 07, 2026RGB-based camouflaged object detection struggles in real-world scenarios where color and texture cues are ambiguous. While hyperspectral image offers a powerful alternative by capturing fine-grained spectral signatures, progress in hyperspectral camouflaged object detection (HCOD) has been critically hampered by the absence of a dedicated, large-scale benchmark. To spur innovation, we introduce HyperCOD, the first challenging benchmark for HCOD. Comprising 350 high-resolution hyperspectral images, It features complex real-world scenarios with minimal objects, intricate shapes, severe occlusions, and dynamic lighting to challenge current models. The advent of foundation models like the Segment Anything Model (SAM) presents a compelling opportunity. To adapt the Segment Anything Model (SAM) for HCOD, we propose HyperSpectral Camouflage-aware SAM (HSC-SAM). HSC-SAM ingeniously reformulates the hyperspectral image by decoupling it into a spatial map fed to SAM's image encoder and a spectral saliency map that serves as an adaptive prompt. This translation effectively bridges the modality gap. Extensive experiments show that HSC-SAM sets a new state-of-the-art on HyperCOD and generalizes robustly to other public HSI datasets. The HyperCOD dataset and our HSC-SAM baseline provide a robust foundation to foster future research in this emerging area.
Curriculum Multi-Task Self-Supervision Improves Lightweight Architectures for Onboard Satellite Hyperspectral Image Segmentation
Sep 16, 2025



Hyperspectral imaging (HSI) captures detailed spectral signatures across hundreds of contiguous bands per pixel, being indispensable for remote sensing applications such as land-cover classification, change detection, and environmental monitoring. Due to the high dimensionality of HSI data and the slow rate of data transfer in satellite-based systems, compact and efficient models are required to support onboard processing and minimize the transmission of redundant or low-value data, e.g. cloud-covered areas. To this end, we introduce a novel curriculum multi-task self-supervised learning (CMTSSL) framework designed for lightweight architectures for HSI analysis. CMTSSL integrates masked image modeling with decoupled spatial and spectral jigsaw puzzle solving, guided by a curriculum learning strategy that progressively increases data complexity during self-supervision. This enables the encoder to jointly capture fine-grained spectral continuity, spatial structure, and global semantic features. Unlike prior dual-task SSL methods, CMTSSL simultaneously addresses spatial and spectral reasoning within a unified and computationally efficient design, being particularly suitable for training lightweight models for onboard satellite deployment. We validate our approach on four public benchmark datasets, demonstrating consistent gains in downstream segmentation tasks, using architectures that are over 16,000x lighter than some state-of-the-art models. These results highlight the potential of CMTSSL in generalizable representation learning with lightweight architectures for real-world HSI applications. Our code is publicly available at https://github.com/hugocarlesso/CMTSSL.
SpecAware: A Spectral-Content Aware Foundation Model for Unifying Multi-Sensor Learning in Hyperspectral Remote Sensing Mapping
Oct 31, 2025



Hyperspectral imaging (HSI) is a vital tool for fine-grained land-use and land-cover (LULC) mapping. However, the inherent heterogeneity of HSI data has long posed a major barrier to developing generalized models via joint training. Although HSI foundation models have shown promise for different downstream tasks, the existing approaches typically overlook the critical guiding role of sensor meta-attributes, and struggle with multi-sensor training, limiting their transferability. To address these challenges, we propose SpecAware, which is a novel hyperspectral spectral-content aware foundation model for unifying multi-sensor learning for HSI mapping. We also constructed the Hyper-400K dataset to facilitate this research, which is a new large-scale, high-quality benchmark dataset with over 400k image patches from diverse airborne AVIRIS sensors. The core of SpecAware is a two-step hypernetwork-driven encoding process for HSI data. Firstly, we designed a meta-content aware module to generate a unique conditional input for each HSI patch, tailored to each spectral band of every sample by fusing the sensor meta-attributes and its own image content. Secondly, we designed the HyperEmbedding module, where a sample-conditioned hypernetwork dynamically generates a pair of matrix factors for channel-wise encoding, consisting of adaptive spatial pattern extraction and latent semantic feature re-projection. Thus, SpecAware gains the ability to perceive and interpret spatial-spectral features across diverse scenes and sensors. This, in turn, allows SpecAware to adaptively process a variable number of spectral channels, establishing a unified framework for joint pre-training. Extensive experiments on six datasets demonstrate that SpecAware can learn superior feature representations, excelling in land-cover semantic segmentation classification, change detection, and scene classification.
Addressing Annotation Scarcity in Hyperspectral Brain Image Segmentation with Unsupervised Domain Adaptation
Aug 23, 2025This work presents a novel deep learning framework for segmenting cerebral vasculature in hyperspectral brain images. We address the critical challenge of severe label scarcity, which impedes conventional supervised training. Our approach utilizes a novel unsupervised domain adaptation methodology, using a small, expert-annotated ground truth alongside unlabeled data. Quantitative and qualitative evaluations confirm that our method significantly outperforms existing state-of-the-art approaches, demonstrating the efficacy of domain adaptation for label-scarce biomedical imaging tasks.
SAMSA: Segment Anything Model Enhanced with Spectral Angles for Hyperspectral Interactive Medical Image Segmentation
Jul 31, 2025Hyperspectral imaging (HSI) provides rich spectral information for medical imaging, yet encounters significant challenges due to data limitations and hardware variations. We introduce SAMSA, a novel interactive segmentation framework that combines an RGB foundation model with spectral analysis. SAMSA efficiently utilizes user clicks to guide both RGB segmentation and spectral similarity computations. The method addresses key limitations in HSI segmentation through a unique spectral feature fusion strategy that operates independently of spectral band count and resolution. Performance evaluation on publicly available datasets has shown 81.0% 1-click and 93.4% 5-click DICE on a neurosurgical and 81.1% 1-click and 89.2% 5-click DICE on an intraoperative porcine hyperspectral dataset. Experimental results demonstrate SAMSA's effectiveness in few-shot and zero-shot learning scenarios and using minimal training examples. Our approach enables seamless integration of datasets with different spectral characteristics, providing a flexible framework for hyperspectral medical image analysis.
Omni-Fusion of Spatial and Spectral for Hyperspectral Image Segmentation
Jul 09, 2025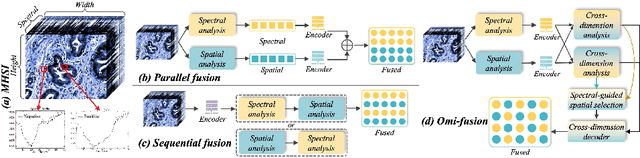
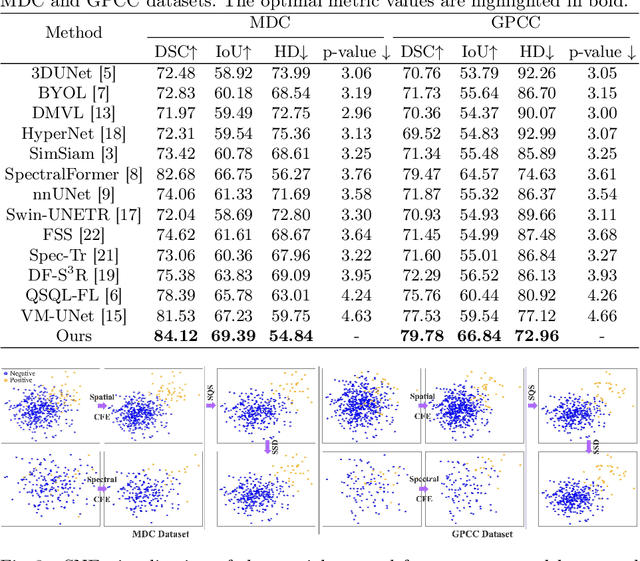
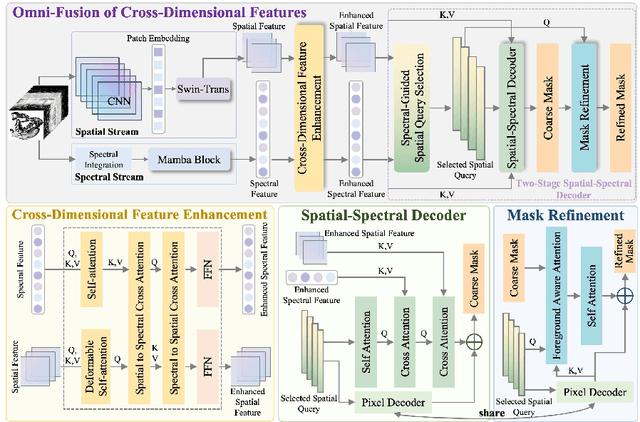
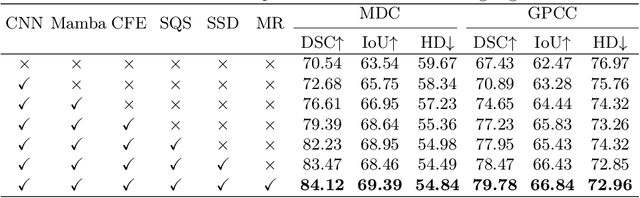
Medical Hyperspectral Imaging (MHSI) has emerged as a promising tool for enhanced disease diagnosis, particularly in computational pathology, offering rich spectral information that aids in identifying subtle biochemical properties of tissues. Despite these advantages, effectively fusing both spatial-dimensional and spectral-dimensional information from MHSIs remains challenging due to its high dimensionality and spectral redundancy inherent characteristics. To solve the above challenges, we propose a novel spatial-spectral omni-fusion network for hyperspectral image segmentation, named as Omni-Fuse. Here, we introduce abundant cross-dimensional feature fusion operations, including a cross-dimensional enhancement module that refines both spatial and spectral features through bidirectional attention mechanisms, a spectral-guided spatial query selection to select the most spectral-related spatial feature as the query, and a two-stage cross-dimensional decoder which dynamically guide the model to focus on the selected spatial query. Despite of numerous attention blocks, Omni-Fuse remains efficient in execution. Experiments on two microscopic hyperspectral image datasets show that our approach can significantly improve the segmentation performance compared with the state-of-the-art methods, with over 5.73 percent improvement in DSC. Code available at: https://github.com/DeepMed-Lab-ECNU/Omni-Fuse.
Broadband Near-Infrared Compressive Spectral Imaging System with Reflective Structure
Aug 20, 2025



Near-infrared (NIR) hyperspectral imaging has become a critical tool in modern analytical science. However, conventional NIR hyperspectral imaging systems face challenges including high cost, bulky instrumentation, and inefficient data collection. In this work, we demonstrate a broadband NIR compressive spectral imaging system that is capable of capturing hyperspectral data covering a broad spectral bandwidth ranging from 700 to 1600 nm. By segmenting wavelengths and designing specialized optical components, our design overcomes hardware spectral limitations to capture broadband data, while the reflective optical structure makes the system compact. This approach provides a novel technical solution for NIR hyperspectral imaging.
FusionSort: Enhanced Cluttered Waste Segmentation with Advanced Decoding and Comprehensive Modality Optimization
Aug 27, 2025



In the realm of waste management, automating the sorting process for non-biodegradable materials presents considerable challenges due to the complexity and variability of waste streams. To address these challenges, we introduce an enhanced neural architecture that builds upon an existing Encoder-Decoder structure to improve the accuracy and efficiency of waste sorting systems. Our model integrates several key innovations: a Comprehensive Attention Block within the decoder, which refines feature representations by combining convolutional and upsampling operations. In parallel, we utilize attention through the Mamba architecture, providing an additional performance boost. We also introduce a Data Fusion Block that fuses images with more than three channels. To achieve this, we apply PCA transformation to reduce the dimensionality while retaining the maximum variance and essential information across three dimensions, which are then used for further processing. We evaluated the model on RGB, hyperspectral, multispectral, and a combination of RGB and hyperspectral data. The results demonstrate that our approach outperforms existing methods by a significant margin.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge